Transposable Elements in Human Cancer
Dr Alfred Grech & Dr Stephen West
ABSTRACT
Transposable elements are repetitive DNA sequences consisting of RNA transposons, DNA transposons, and endogenous retroviruses. Repetitive sequences cover more than two-thirds of the genome in humans and transposable elements comprise the majority of the repetitive sequences (approximately 50% of the human genome).
Transposable elements can transpose (i.e. can jump) and cause havoc. Indeed, research is showing that their deregulation impinges on the stability of the genome and on the regulation of transcription and that of non-coding RNA. This leads to carcinogenesis and to cancer progression. These insights are furthering their research to discover novel targets for theranostic applications in cancer.
NTRODUCTION
A ‘transposable element’ (TE) is a DNA sequence that can move and change its position (transpose) within the same chromosome or from one chromosome to another. These mobile DNA sequences are also called ‘mobile elements’. It was the geneticist Barbara McClintock back in 1948 that discovered these elements in her seminal work on the mosaic colouration in maize. Her theory met a lot of scepticism in the field, and had to wait years before her work was recognized. Indeed, it was in 1983 when she was awarded the Nobel prize in Physiology/Medicine. Since then, similar mobile elements have been discovered in mammalian genomes, including that of humans. Moreover, they have also been found in almost all living species, including bacteria. The presence of active mobile DNAs in humans was only appreciated in 1988, when research showed that a TE was responsible for Haemophilia A. Since then, several genetic diseases have been discovered to be mediated by TEs.
DNA transposons and retrotransposons (also called retro-elements) are the two main classes of transposable elements. This classification is based on whether an RNA intermediate is involved during transposition. DNA TE hop by a “cut-and-paste” mechanism, while retrotransposons replicate via a “copy-and-paste” mechanism. This essay will focus on retrotransposons and specifically, on LINE-1 (long interspersed nuclear element-1, also called L1), which are the most well-studied retrotransposons.
The human genome harbours over 500,000 copies of LINE-1 elements and it is estimated that they represent 18% of the genetic code. In the human somatic cell, the majority of LINE-1 elements are not active. In fact, it has been estimated that there are only 80 to 100 LINE[1]1 elements that are ‘hot’, so defined because they are capable of propagation (‘retrotransposon competent’ L1). Scientists call these subsets of LINE-1 elements as ‘genetic parasites’.1
STRUCTURE OF LINE-1 ELEMENTS
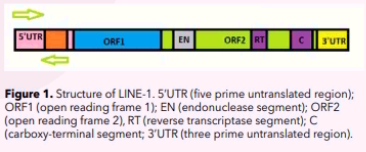
LINE-1 elements are approximately 6 –7 kilobases (kb) in length. Simplistically, a LINE-1 element consists of the following parts:
- a 5’-UTR (five prime untranslated region) which is rich in cytosine-phosphate-guanine (CpG) – this region has two internal promoters, one is sense and the other is antisense;
- two open reading frames (ORFs) which do not overlap, with ORF1 being 1 kb long and ORF2 being 3.8 kb long;
- a 3’-UTR (206 nucleotides) with a poly(A) tail
THE PROCESS OF L-1 RETROTRANSPOSITION
Briefly, the process of L-1 retrotransposition consists of the following stages:
- A cycle starts in the nucleus with the disassembly of the nucleosomal and remodeling deacetylase (NuRD) multiprotein complex from the L1 promoter. NuRD is a repressor complex, which together with epigenetic repressor marks (e.g. histone-3 lysine-9 trimethylations (H3K9me3) and histone-3 lysine-20 trimethylations (H3K20me3)) cause L1 promoter to be in a heterochromatic state, the latter being a compact condensed state of the DNA making it inaccessible for transcription. Thus, removal of NuRD and the epigenetic repressor marks, shifts the L1 promoter from its heterochromatic state to a euchromatic one, which is open and uncompacted chromatin, allowing transcription.
- The L-1 DNA sequence is transcribed by RNA polymerase II into the L1 mRNA. The RNA polymerase binds to the sense promoter and starts transcription in a 5’ to 3’ manner.
- L1 mRNA exports to the cytoplasm.
- In the cytoplasm, L1 mRNA is translated. ORF1 frame encodes ORF1 protein (ORF1p) which is 40kDa and has an RNA recognition motif. ORF2 frame encodes ORF2 protein (ORF2p) which is 150kDa and has endonuclease and reverse transcriptase activities.
- In the majority of cases ORF1p and ORF2p proteins preferentially assemble with their own L-1 transcript forming L1 ribonucleoprotein complex (L1 RNP). This preference binding is called cis preference. However, a minority of them show trans-preference. Indeed, they can bind to SINEs (short interspersed nuclear elements) and other mRNAs to form RNPs.
- From the cytoplasm the L1 RNP complex imports into the nucleus.
- Reverse transcription of the L1 mRNA occurs producing the L1 complementary DNA (L1 cDNA). The endonuclease activity of ORFp targets a DNA sequence (5=-TTTTAA-3=), nicks it and the L1 element is inserted. This process is called ‘target-site primed reverse transcription’ (TPRT).2
MECHANISMS THAT SILENCE TEs
One of the somatic mechanisms that the host cell uses to inhibit the expression of L1 and its transposition is ‘epigenetic repression’. Epigenetics refers to reversible chemical mechanisms that affect gene expression without altering the DNA sequence. Some of these epigenetic mechanisms affect the L1 5’UTR and notably include DNA hypermethylation, repressive histone modifications (H3K9me3, H3K20me3) and recruitment of the nucleosome remodeling and deacetylase (NuRD) complex. These changes cause the chromatin structure to condense into the ‘inaccessible’ heterochromatin and so transcription cannot occur. Another epigenetic repressive mechanism is that by micro-RNAs. Micro-RNA-induced L1 silencing is mediated by the micro-RNA binding directly to the L1 mRNA, which is then degraded. Specifically one such micro-RNA is microRNA-128.3
It is obvious that L1 elements like other TEs can cause havoc when unleashed. That is why the host response to limit the harmful effects of these TEs is a multilayered one directed at the various stages of their life cycle. Below are some of the known processes that the mammalian somatic cell has developed to repress their activity. Many of the processes ‘cross talk’ with each other and form complicated networks involving several stages and molecules. These repressive mechanisms have been found to be deregulated in cancer cells or in cells that are exposed to environmental carcinogens, thus allowing L1 retrotransposition and propagation.
Besides micro-RNAs, L1 mRNA is also degraded or suppressed by small interfering RNAs (siRNAs) and piwi[1]interacting RNAs (piRNAs).4
Another system that is triggered when L1 is activated is that involving the Apolipoprotein B (apo B)-editing catalytic (APOBEC) gene family. Its action is to catalyse the deamination of cytosine to uracil in L1 mRNA.5 These cytosine-to-uracil point mutations are then recognized and L1 mRNA is degraded by endonucleases or RNA degrading enzymes.
Another mechanism is the preferential insertions of L1 sequences to gene-poor regions like those found in chromosome 13.6 Chromosome 13 (like chromosome 18, 21, and Y) is a gene-poor chromosome. Thus, new insertions of L1s will unlikely disrupt any gene loci.
L1 mRNA and its proteins can also be sequestered into storage centres called stress granules (SG). These granules are cytosolic membraneless compartments where stalled translation complexes are stored. Being thus sequestered, the RNP is prevented from entering into the nucleus and complete the retrotransposition cycle.7 Subsequently, the L1 mRNA and its proteins are degraded in cytoplasmic membraneless P-bodies (PB) or autophagosomes. PBs and SGs then ‘dock’ and shuttle the LI mRNA and its proteins.8,9
If by any chance the above mechanisms fail and LI RNP manages to enter into the nucleus, other inhibitory mechanisms come into play. An important one involves the highly conserved ERCC1/XPF complex (excision repair cross-complementation group 1/ xeroderma pigmentosum complementation group F), which has endonuclease activity and is involved in various DNA repair pathways to keep the genome stable. Specifically, here, during the TPRT process the complex recognizes the cDNA and removes it, restoring the DNA sequence.10
LINE-1 MOBILISATION AND TUMORIGENESIS
Transposable elements shaped and still are shaping our genome. However, their jumping around can cause genomic instability and cause disease, including cancer.
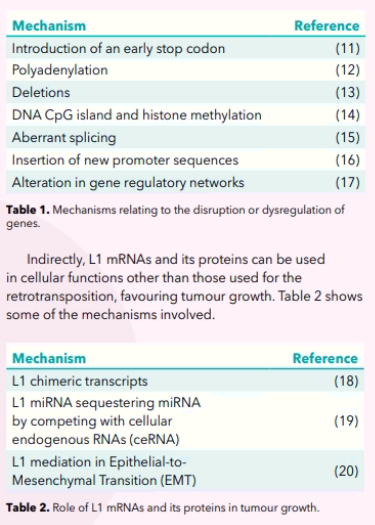
Several mechanisms have been proposed for the role of L1 in tumorigenesis. L1 can be involved directly or indirectly. Briefly, when they insert their sequence through retrotransposition, L1 directly can disrupt or dysregulate the structure and function of genes by the following mechanisms (Table 1).
A biomarker is of benefit if it can provide useful information in the diagnosis (especially early detection) and/or prognosis and/or monitoring of the effect of treatment. It can also help in the stratification of patients and hence in their tailored treatment. The extensive accumulating research on L1 is showing that it has the potential to be such a biomarker in cancer. Specifically, the following are being proposed as L1 biomarkers: L1-ORF1, L1-ORF2, L1- mRNA, L1-methylation status and L1-DNA.
Various cancers release cancer cells into the circulation. Circulating L1-DNA from these circulating cancer cells has been proposed by Sunami et al.21 as a biomarker in breast cancer. They found that L1-DNA was higher in breast cancer patients and could be useful in detecting early-stage breast cancer. Also, they observed a correlation between LI-DNA copy number and tumour size. Another study22 also showed that circulating L1-DNA level was associated with tumour size.
Other feasible L1-biomarkers are L1-mRNA and its proteins (L1-ORF1 and L1-ORF2). L1-mRNA can be profiled in tumours. More importantly, using tissue specific markers, L1-mRNA can be traced back to the tissue releasing L1,23 thus serving as an indirect approach to determine the tissue of origin. Over-expression of L1-ORF2 protein was reported in colon, lung, breast and prostate cancer of epithelial phenotypes.24 Similarly, expression of L1-ORF1 protein was found in several invasive tumours25 and was associated with a poor prognosis.
It has been shown that L1-hypomethylation is linked with a poor prognosis. For example L1-hypomethylation in tissue samples of ovarian cancer,26 colon cancer,27 glioblastoma multiforme28 and hepatocellular carcinoma29 was associated with a lower 5-year survival rate. L1- hypomethylation can also be used to stratify patients to treatment.30
LINE-1 AS A POTENTIAL THERAPEUTIC TARGET IN CANCER
L1 life cycle offers a plethora of molecules that can be targeted to inhibit its propagation and thus inhibit L1- mediated mutagenesis.
Line-1’s ORF2 reading frame encodes the protein ORF2p which has endonuclease and reverse transcriptase activities. L-1 inhibition by reverse transcriptase inhibitors can thus be a potential treatment in cancer by targeting reverse transcriptase activity.
In 2005, Sciamana et al.31 used two reverse transcriptase inhibitors (nevirapine and efavirenz) to slow down the growth proliferation of cells in cell lines of prostatic carcinoma and malignant melanoma.
n 2010, Carini et al.32 also showed the effectiveness of another reverse transcriptase inhibitor, abacavir, on the proliferative growth of prostatic cancer.
In 2014, the FAVE clinical study33 showed the efficacy of efavirenz in metastatic castration-resistant cancer of the prostate. In a follow-up phase 1 clinical trial (Identifier: NCT01878890) called ESCALE, efavirenz was tested on patients with solid tumours and Non-Hodgkin lymphoma in higher doses than those used in the FAVE study. This was done to test doses above 600mg daily in order to establish the maximum tolerated therapeutic dose.
Translational medical researchers have always looked into photochemicals for their therapeutic use. This is because they function in numerous biological pathways and besides, these usually have a low toxic profile. Nishikawa et al.34 screened various phytochemicals and found that capsaicin has reverse transcriptase inhibition properties and it suppresses L1 retrotransposition. They thus propose that one way in which capsaicin may defeat the progression of tumorigenesis is by inhibiting L1- mediated mutagenesis. They also propose further studies to investigate capsaicin and related compounds called capsaicinoids in the quest to prevent and treat cancer. RNA-based treatments are also gaining momentum.
Another pathway of treating cancer based on L1 silencing might be the use of micro-RNA mimics. It has already been Vol 21 2022 • Issue 02 thesynapse.net • 17 pointed out that miR-128 represses L1 activity. Idica et al.35 found that miR-128 binds directly to L1 ORF2 RNA thus repressing its transposition.
CONCLUSION The ubiquitous transposable elements have been underappreciated because they have been labelled as unimportant and also, because of their sheer high number of copies and variants, created technical challenges. These obstacles have now been overturned. Unfaltering research and new methodical platforms are showing that dysregulated expression of TE is a characteristic of cancer and possibly plays an important role in tumour initiation and progression. As more ongoing translational research on TEs unfolds, one hopes that it will soon be applied in oncology clinical practice in the field of diagnostic and prognostic biomarkers and also, as potential therapeutic targets.
REFERENCES
- Montoya-Durango DE, Ramos KA, Bojang P, et al. LINE-1 silencing by retinoblastoma proteins is effected through the nucleosomal and remodeling deacetylase multiprotein complex. BMC Cancer 2016;16:38.
- Luan DD, Korman MH, Jakubczak JL, et al. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 1993;72(4):595-605.
- Hamdorf M, Idica A, Zisoulis DG, et al. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nature Structural & Molecular Biology 2015;22(10):824-31.
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell 2009;136(4):763-76.
- Ikeda T, Abd El Galil KH, Tokunaga K, et al. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Research 2011;39(13):5538-54.
- Bojang P, Jr., Anderton MJ, Roberts RA, et al. De novo LINE-1 retrotransposition in HepG2 cells preferentially targets gene poor regions of chromosome 13. Genomics 2014;104(2):96-104.
- Li X, Zhang J, Jia R, et al. The MOV10 helicase inhibits LINE-1 mobility. The Journal Of Biological Chemistry 2013;288(29):21148-60.
- Hu S, Li J, Xu F, et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genetics 2015;11(7):e1005367.
- Guo H, Chitiprolu M, Gagnon D, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nature Communications 2014;5(1):5276.
- Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair 2008;7(6):983-9.
- Narita N, Nishio H, Kitoh Y, et al. Insertion of a 5’ truncated L1 element into the 3’ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. The Journal Of Clinical Investigation 1993;91(5):1862-7.
- Iskow RC, McCabe MT, Mills RE, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 2010;141(7):1253-61.
- Samuelov L, Fuchs-Telem D, Sarig O, et al. An exceptional mutational event leading to Chanarin-Dorfman syndrome in a large consanguineous family. Br J Dermatol 2011;164(6):1390-2.
- Liu N, Lee CH, Swigut T, et al. Selective silencing of euchromatic L1s revealed by genome-wide screens for L1 regulators. Nature 2018;553(7687):228-32.
- Kazazian HH, Jr., Wong C, Youssoufian H, et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 1988;332(6160):164-6.
- Muotri AR, Chu VT, Marchetto MC, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005;435(7044):903-10.
- Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. The EMBO Journal 1996;15(3):630-9.
- Ohms S, Lee SH, Rangasamy D. LINE-1 retrotransposons and let[1]7 miRNA: partners in the pathogenesis of cancer? Frontiers in Genetics 2014;5:338.
- Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives.Tumour Biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 2015;36(2):479-88.
- Reyes-Reyes EM, Aispuro I, Tavera-Garcia MA, et al. LINE-1 couples EMT programming with acquisition of oncogenic phenotypes in human bronchial epithelial cells. Oncotarget 2017;8(61):103828-42.
- Sunami E, Vu AT, Nguyen SL, et al. Quantification of LINE1 in circulating DNA as a molecular biomarker of breast cancer. Annals of the New York Academy of Sciences 2008;1137:171-4.
- Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Communications 2011;2:180.
- De Luca C, Guadagni F, Sinibaldi-Vallebona P, et al. Enhanced expression of LINE-1-encoded ORF2 protein in early stages of colon and prostate transformation. Oncotarget 2016;7(4):4048-61.
- Feng F, Lu YY, Zhang F, et al. Long interspersed nuclear element ORF-1 protein promotes proliferation and resistance to chemotherapy in hepatocellular carcinoma. World Journal of Gastroenterology 2013;19(7):1068-78.
- Harris CR, Normart R, Yang Q, et al. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes & Cancer 2010;1(2):115-24.
- Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. International journal of gynecological cancer 2008;18(4):711-7.
- Wang BG, Huang HY, Chen YC, et al. Increased plasma DNA integrity in cancer patients. Cancer Research 2003;63(14):3966-8.
- Ohka F, Natsume A, Motomura K, et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One 2011;6(8):e23332.
- Harada K, Baba Y, Ishimoto T, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Annals of Surgical Oncology 2015;22(4):1280-7.
- Kawakami K, Matsunoki A, Kaneko M, et al. Long interspersed nuclear element-1 hypomethylation is a potential biomarker for the prediction of response to oral fluoropyrimidines in microsatellite stable and CpG island methylator phenotype-negative colorectal cancer. Cancer Science 2011;102(1):166-74.
- Sciamanna I, Landriscina M, Pittoggi C, et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 2005;24(24):3923-31.
- Carlini F, Ridolfi B, Molinari A, et al. The reverse transcription inhibitor abacavir shows anticancer activity in prostate cancer cell lines. PloS one 2010;5(12): e14221.
- Houédé N, Pulido M, Mourey L, et al. A phase II trial evaluating the efficacy and safety of efavirenz in metastatic castration-resistant prostate cancer. Oncologist 2014;19(12):1227-8.
- Nishikawa Y, Nakayama R, Obika S, et al. Inhibition of LINE-1 Retrotransposition by Capsaicin. International Journal of Molecular Sciences 2018;19(10).
- Idica A, Sevrioukov EA, Zisoulis DG, et al. MicroRNA miR-128 represses LINE-1 (L1) retrotransposition by down-regulating the nuclear import factor TNPO1. The Journal of Biological Chemistry 2017;292(50):20494-508.

