The genomics of type 2 diabetes – the Maltese contribution
By Nikolai P Pace, JosanneVassallo& Alex E Felice
Abstract
The genomic revolution has transformed the type 2 diabetes genetic landscape, with many loci reported to have association with disease risk. Such loci exert a weak to modest effect size and are as yet of limited use in risk prediction studies. They also fail to account for robust genotype-phenotype associations. This study aims to address these issues by looking at the quantitative summation of risk alleles and the use of special populations to better define risk and phenotypic associations.
Introduction
Diabetes Mellitus type 2 (T2DM) is a common, etiologically complex chronic disorder that arises from the elaborate interplay between lifestyle factors and various genetic elements. It represents one of the most widespread public health problems in both the developed and developing world. Commonly, the impaired regulation of glucose homeostasis that leads to type 2 diabetes is associated with a cluster of cardio-metabolic risk factors – including abdominal obesity, hypertension andatherogenicdyslipidaemia, which is a chronic subclinical proinflammatory state. These factors combine to determine the development and progression of both microvascular and macrovascular disease that carry a substantialsocio-economic burden globally. Central to the development of obesity, insulin resistance and type 2 diabetes is a distinct lifestyle component defined by the overconsumption of calorie-dense food and a lack of physical exercise.
Background
The study of complex disease genetics has undergone a massive revolution over the past decade, driven largely by the completion of the Human Genome and the International HapMap projects.1 These have provided researchers with innovative insight into population-specific patterns of common human genetic variation. Simultaneously, technological advances have led to the development of DNA microarray platforms. These commercially available ‘DNA chips’ utilize known linkage disequilibrium patterns to genotype tag polymorphisms and enable the imputation of genotypes at linked loci. Ongoing developments in second-generation sequencing technology have enabled the processing of millions of sequence reads in parallel. These have continued to drive down the cost and timeframes necessary to obtain whole genome and transcriptome datasets which are vital to understanding complex diseases.
Candidate Gene or Genome-Wide Association Studies?
The search for T2DM susceptibility loci has been fraught with complications. The disease is genetically heterogeneous, and strong gene-environment interactions influence its onset and progression. Furthermore, the diagnosis of T2DM hinges on the identification of elevated fasting or post-prandial blood glucose levels that lead to metabolic and microvascular complications. It is highly likely that such a diagnosis masks a tremendous amount of clinical heterogeneity, and that the genetic heterogeneity behind diabetes is as great as its clinical heterogeneity.2 Hypothesis-driven candidate gene studies have identified a few genetic variants associated with T2DM. These include common variants in peroxisome proliferator-activated receptor γ (PPARy)3, the KCNJ11 potassium channel4 and the transcription factor TCF7L2.5 Such candidate gene studies are based on selection of loci with a known or inferred biological function which may predispose to disease or the observed phenotype.6Nevertheless such a priori assumption inherently limits the capacity of candidate gene studies to identify pathways or loci playing previously unsuspected roles in the etiology or pathogenesis of complex diseases.
The genomics revolution and the subsequent introduction of Genome-Wide Association Studies (GWAS) in 2007has lead to a massive upsurge in the number of T2DM-associated loci. To date, the online catalog of published Genome-Wide Association Studies (www.genome.gov/gwastudies/) lists 43 GWAS that have investigated T2DM as a disease of which 8 GWAS have also investigated fasting blood glucose as a quantitative trait. As larger collections continue to be assembled for meta-analysis by international consortia, new associations with T2DM, fasting blood glucose and fasting insulin continually emerge.7
GWAS – a failed promise?
Hundreds of variants identified through GWAS have been associated with complex traits or diseases, including T2DM. In some cases, the identified variants have provided valuable insight into the genetic architecture of disease. However, GWAS-identified variants have a low effect size and consequently confer only slight increments in risk and account for a minor proportion of familial clustering.8GWAS-identified associations are estimated to explain only around 10% of the heritability of T2DM. They fail to account for epistasis, epigenetic changes and gene-environment interactions. They are largely restricted to populations of European descent and carry poor discriminatory capacity that limits their use in risk prediction. Furthermore, the transition from genotype to functional physiology is not straightforward. While polymorphisms in promoter or exon regions have predictable biological effects, variants in deep intronic or intergenic regions have probable regulatory roles. GWAS also utilise large cohorts of patients that are often heterogeneous or poorly characterised, thus limiting the association of genotype data with clinical or biochemical phenotypes.
Type 2 Diabetes Genetics and the Maltese Population
The Maltese population has a high predisposition for T2DM. The earliest evidence concerning interest in diabetes in Malta dates back to 1698.9The period following the Second World War witnessed an improvement in socio-economic conditions which, despite the introduction of oral hypoglycemic agents, was accompanied by a marked increase in diabetes-specific mortality. The estimated national prevalence for T2DM in 2013 stands at 10.14% and is expected to rise to 11.44% by 2035.10
Two studies have investigated the genetics of T2DM in the Maltese population.11,12The aims are listed hereunder.
- Further define the genetic interplay between a carefully selected panel of genes from metabolic and inflammatory pathways and the risk of developing type 2 diabetes in adulthood.
- To relate the association of defined genetic profiles with biological and clinical endpoints and monocyte gene expression profiles to type 2 diabetes.
These studies were innovative in several key points. A total of 42 genetic variants were selected for genotyping from genes in several metabolic and/or inflammatory pathways that impact on glucose regulation, lipid metabolism, inflammation and insulin resistance. The genetics of the disease was investigated using different comparative populations, including 600 Maltese and 200 Libyan T2DM cases. The Maltese T2DM cases were further subdivided into carefully defined subpopulations of treatment-naïve and high-morbidity groups. A unique collection of two hundred cord blood DNA samples obtained from the Malta BioBank (www.um.edu.mt/biobank/home) was selected as the control reference population. These were carefully selected to be of Maltese ethnicity and to exclude close siblings.
The salient findings of these investigations are the following:
- The quantitative summation of risk alleles has been associated with significant increases in the risk of developing T2DM (figure1).Nevertheless, when individual cases are considered, risk polymorphisms exerted only a slight-to-modest effect on disease risk. Consequently, risk alleles studied in isolation contribute minimally to disease risk prediction. Equally interesting is the high odds ratios for selected risk alleles reported in thesestudies, in particular for a common variant in the beta-2 adrenergic receptor (ADRβ2).
- The recruitment of a carefully-characterised treatment-naïve cohort is a powerful tool in the identification of genotype-phenotype associations. The clinical, anthropometric and biochemical risk factors were, as much as possible and to our knowledge, unaltered by drugs and/or lifestyle change. Too often, published genotype-phenotype associations included larger numbers of less well-characterized subjects that were clinically heterogeneous. Such associations have doubtful pathophysiological relevance, lacked reproducibility or were replicated with disconcordant findings.Drugs commonly used in the management of T2DM, including metformin, statins and aspirin, exert their effects on multiple tissue types through various signaling pathways that could rapidly alter the systemic metabolic and inflammatory profile. Of note, the authors have identifiedan association between a promoter polymorphism in the melatonin-receptor 1 beta gene (MTNR1β) and total cholesterol and LDL-C levelswhich has also been reported in other ethnic groups.13Other significant genotype-phenotype associations which have been identified include promoter polymorphism in Interleukin-6 with insulin resistance index (HOMA-IR), and an insulin growth factor 1 variant (IGF1) with body weight.
- Despite the increased effort to recruit treatment-naïve subjects and the aforementioned findings, genotype-phenotype associations still remain considerably deficient. This strongly suggests that while genetic factors contribute to disease risk and onset, the end clinical phenotype is highly variable and is influenced by complex non-genetic factors.
- Microarray gene expression analysis of peripheral blood leukocytes in T2DM provides an intriguing insight into their role ininflammation and the development and progression of diabetic complications. While there is no clear direct relationship between genetic risk scores and gene expression profiles, bioinformatic approaches have identifieddifferentially expressed genes that have established roles in inflammation, oxidative damage and leukocyte activation.
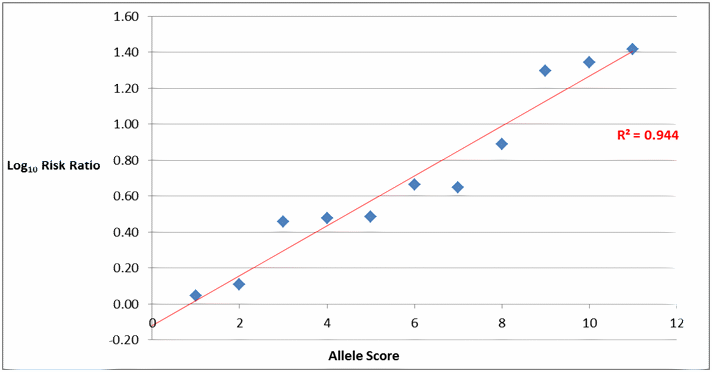
Figure 1:Log10 risk ratio of T2DM in a studymeta-analysis vs. increasing number of risk alleles.An unweighted genetic score was constructed by the simple count of risk alleles. This method assumed that each allele exerts equal and additive effects that can be summed. Genetic risk scores identify a gradient of genetic risk in the T2DM population, with individuals at the high end of the genetic risk score distribution having a markedly higher individual risk than those at the lower end of the genetic risk score distribution. Th e predictive nature of genetic risk scores in this investigation is limited by the cross-sectional study design.
References
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007 Oct 18;449(7164):851–61.
- Permutt MA, Wasson J, Cox N. Genetic epidemiology of diabetes. J Clin Invest. 2005 Jun;115(6):1431–9.
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000 Sep;26(1):76–80.
- Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, Hitman G, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003 Feb;52(2):568–72.
- Winckler W, Weedon MN, Graham RR, McCarroll SA, Purcell S, Almgren P, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007 Mar;56(3):685–93.
- Barroso I, Luan J, Middelberg RPS, Harding A-H, Franks PW, Jakes RW, et al. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. Plos Biol. 2003 Oct;1(1):E20.
- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University. Online Mendelian Inheritance in Man, OMIM®. [Internet]. Available from: http://omim.org/
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009 Oct 8;461(7265):747–53.
- C Savona Ventura. The History of Diabetes Mellitus – A Maltese perspective [Internet]. Malta; 2002. Available from: http://staff.um.edu.mt/csav1/books/diabetes_hist.pdf
- International Diabetes Federation. IDF Diabetes Atlas, 6th edn [Internet]. 2013. Available from: http://www.idf.org/diabetesatlas
- Al Ashtar AA. Molecular SNPlotypesTM with Common Alleles Reflects Expression Profile in Diabetes Mellitus Type 2 [PhD Thesis]. University of Malta; 2008.
- Pace NP. Does summation of alleles account for genetic risk and genotype-phenotype association in Type 2 Diabetes Mellitus? [PhD Thesis]. University of Malta; 2013.
- Ling Y, Li X, Gu Q, Chen H, Lu D, Gao X. A common polymorphism rs3781637 in MTNR1B is associated with type 2 diabetes and lipids levels in Han Chinese individuals. Cardiovasc Diabetol. 2011 Apr 6;10(1):27.
Acknowledgements
The research work disclosed in this article was funded by the Strategic Educational Pathways Scholarship Scheme – STEPS (Malta) and by institutional funds of the University of Malta.The STEPS scholarship is part-financed by the European Union – European Social Fund (ESF) under Operational Programme II- Cohesion Policy 2007-2013, “Empowering people for More Jobs and a Better Quality of Life”
We acknowledge the invaluable contribution of all the staff and patients at the Diabetes and Endocrine Clinic, Mater Dei Hospital and to the family medicine practitioners who helped in the recruitment of the study cohorts for this project.

