HERVs, Transposons and Human Diseases – Part II
by Alfred Grech & Sandra Baldacchino
The Host Cellular Response To Transposable Elements
It is obvious that TEs can have a neutral, good or bad effect on the genome when they transpose. In the short term TEs can cause havoc when unleashed. The host response to limit the harmful effects of TEs is a multi-layered one directed at the various stages of the life cycle of TEs.1 Table 7 below hints at some of the known processes that the mammalian cell has developed to repress TE activity. Many of the processes ‘cross talk’ with each other and form complicated networks involving several stages and molecules. It is not the intention of this paper to go into these in detail but the main molecular protagonists are mentioned in the table, for those keen to find out more about the subject.
|
TE Life Cycle Stage |
Main Processes Involved |
Molecules Involved |
|
1) TE Transcription |
Epigenetic modifications |
DNA methylation; histone modifications; proteins that bring about the formation of repressive chromatin states |
|
2) Post-Transcriptional Processing of TE RNAs |
(i) RNA Editing (ii) RNAi
|
(i) RNA editases (ADAR family; APOBEC proteins; Dicer protein; Argonaute proteins; short interfering RNA, microRNA, RNA-induced silencing complex, Piwi-interacting RNA ) |
|
3) Integration of TE Copies |
DNA Repair |
DNA repair enzymes e.g. ERCC1/XPF endonuclease |
Table 7: The Multilayered Host Response to TEs
Epigenetic modifications simply refer to chemical tags that are made to the chromatin template rendering it condensed and so repressive for transcription. Specifically, such epigenetic modifications include (i) DNA-methylation and (ii) modifications to tails of histone proteins. Here, some in depth description of these processes is important in order to understand how by knowing them, prevention and treatment could be devised for some of the medical conditions in which TEs are being implicated in their pathogenesis.
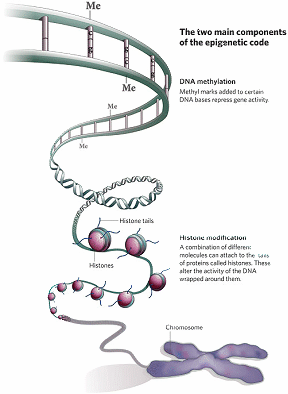
Figure 2: DNA Methylation and Histone Modification are the two main epigenetic modifications in mammals, including humans (Dr Mark Hill, Molecular Development – Epigenetics (2010).
In DNA-methylation, a methyl donor molecule called S-adenosylmethionine donates a methyl group which is covalently added to the carbon-5 position of cytosine within the CpG dinucleotide in the DNA molecule. This reaction is enzyme mediated and is performed by a family of enzymes called DNA methyltransferases (DNMTs).
Histone proteins come together in octamers i.e. 8 molecules, to form the core of nucleosomes around which the DNA double helix wraps. In doing so, they present tails that stick out. These histone tails have amino acids, like lysine and arginine, which can be chemically modified. These chemical modifications of these amino acids on the histone tails affect how the histone proteins interact with DNA, modifying the chromatin template structure and thus act as an epigenetic mechanism. Histone acetylation is one of the best-studied histone modifications. Histone acetyltransferases (HATs) catalyses this acetylation and uses acetyl-coenzyme A as a donor molecule. Histone acetylation occurs largely at lysine residues of the histone H4 and H3, which are core histones of nucleosomes. The level of histone acetylation depends on a balance between the action of HATs and histone deacetylases (HDACs). Another histone modification is that of histone methylation. Histone methylation is harnessed by a family of enzymes called histone methyltransferases (HMTs). On the other hand and to balance the effect of HMTs when needed, histone demethylases (HDMs) remove methyl groups from histone.
Besides these chemical modifications, other proteins are involved, but all may concur to the heterochromatiztion (condensation of chromatin template) of TEs promoters and hence suppress TE expression.
In RNA editing the protein coding sequences of mRNAs are modified without involving splicing.
RNA interference (RNAi) involves several related processes whereby short RNAs (usually 20-30 nucleotides long) target unwanted nucleic acids, like those of viruses invading the cell or those of transposable elements. Indeed, mounting evidence is showing that RNAi is one of the primary defenses that the cell has against viruses and TEs.2-5 In the case of TEs, RNAi targets TE transcripts by cleaving them and then degrading them or else prevents TEs transcription by helping in bringing heterochromatiztion of their sequence inside the host genome (Table 8).
|
1. Cleavage and degradation of targeted RNA |
|
2. Recruiting additional factors that help modify gene expression |
|
3. Epigenetic modification and heterochromatization |
Table 8: The Outcome of RNAi
Some TEs, like non-LTR retrotransposons that cause insertional mutagenesis, can bring about complex chromosomal re-arrangements characterized by γ-H2AX foci on the chromosome. This focus marks where the DNA double-strand breaks to allow the insertion of the TE sequence. Host DNA repair proteins come into action, when they recognize such induced lesions in the DNA and make appropriate repairs. The ERCC1/XPF is one such DNA repair protein complex.6 It is a heterodimer, meaning that it is a molecule with two different subunits. Specifically and functionally, ERCC1/XPF heterodimer has endonuclease properties which help against TE movement during the integration of their life cycle.
Stopping cell division from occurring is also conducive to suppression of retrotransposition.7
Many of the above processes marshalled by the cell to regulate TEs co-evolved as a defence against TEs and have occurred millions of years ago.8
Implicated Physiological/Biological Functions of TE (Including HERVs)
The biological relevance of TE is starting to be understood. As regards HERVs, most of them in the genome have no effect9 because their DNA sequence has been disrupted with ‘frame-shift mutations’, deletions and/or other re-arrangements that render them defective in their three most essential genes (viz. gag, pol, env genes). However, some HERVs could have physiological/biological functions, especially those whose DNA sequence give rise to expressed transcripts and proteins.
(i) Evolution of the host
Evolution works on genetic variation. If the environment changes this creates a selection pressure on a population. Under such circumstances, in evolutionary theory, it is envisaged that only those individuals in a population with the right genetic make-up could adapt because of their genetic diversity, and hence will survive (thus the saying ‘adapt or perish’). TEs including HERVs could have given rise to countless genetic variations10 and this could have helped in the evolution of various species, including our own.
So here one sees a situation where the host gets an advantage from the endogenisation (integration inside the genome) of viral sequences. The opposite situation, that is, that where a virus acquires genes from its hosts is also feasible and indeed, many mammalian viruses have done so during their evolutionary pathway.
(ii) Mammalian reproduction vis-à-vis placental morphogenesis
Sha Mi et al.11 have shown that HERV-W transcribes a gene (specifically its env gene) that encodes a protein called syncytin. The expression of this gene occurs mostly in placental syncytiotrophoblasts. Sha Mi et al. also showed that the expression of syncytin in other cells leads to the fusion of cells forming giant synctia. Their research work clearly shows that HERV-W is thus important in the generation of the human placenta.
(iii) Acquired immunity in vertebrates
A fusion protein is formed when two or more genes are joined together. Antibodies are such fusion proteins which are produced by the process of VDJ recombination. The latter is a complex process that involves cleaving and rearranging DNA sequences that code for the antigen receptors of immunoglobulins. The DNA rearrangements result in various combinations that contribute to a wide diversity of possible antibodies, which can even range into thousands. It is believed that this VDJ machinery evolved from transposons (the transposon hypothesis of VDJ recombination).12 RAG1 and RAG2 (for recombination-activating genes) are two essential genes for VDJ recombination and it is proposed that the DNA now coding RAG1 and RAG2 was once a mobile element.
(iv) Modulation of gene expression
Some definitions (Table 9 below) are appropriate here in order to understand the roles played by some TEs in modulating gene expression.
|
Promoter |
DNA sequence of a gene to which RNA polymerase binds to start transcription. |
|
Enhancer |
DNA sequence that has a transcriptional regulatory function; it can be can be located at a site remote from the promoter. |
|
Alternative splicing |
is the generation of diverse mRNAs by varying the pattern of pre-mRNA splicing. |
|
RNA editing |
RNA processing events that alter the protein coding sequences of mRNAs; it does not involve splicing. |
|
Polyadenylation |
is the process of adding a poly-A tail to a pre-mRNA. It signals the end of transcription. Most mRNAs in eukaryotes are polyadenylated, and the poly-A tails regulate both translation and mRNA stability.13 |
Table 9: Some Definitions to Understand Modulation of Gene Expression of Some TEs
Genes have a split structure in which segments of coding sequence (called exons) are separated by noncoding sequences (intervening sequences, or introns). When a gene is transcribed a pre-mRNA (also called primary RNA transcript) is produced. In a process called splicing, introns are removed and the exons are joined together, and a mature mRNA is formed. Splicing leads to the synthesis of different homologs from the same primary RNA transcript, since exons of the latter can be shuffled around before being joined together. Homologs imply molecules that have the same origin but differ in function.14
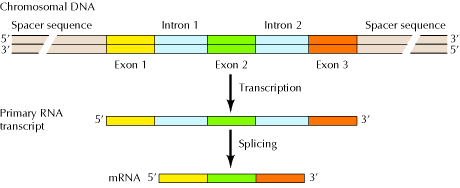
Figure 3: The Structure of Eukaryotic Genes15
Exaptation is the process whereby relics of TEs acquire a regulatory function. For example, HERVs have their own transcriptional promoters, regulators and enhancers in their Long Terminal Repeats (LTRs). Thus these sequences if not disrupted can and do initiate transcription of their gag, pol and env genes, but not only. They also start transcription of neighbouring genes. HERVs can also signal splicing generating diverse transcripts (mRNA) of neighbouring cellular genes. Modulations of genes have also been demonstrated by Alu elements. This occurs at the post-transcriptional level in three ways, (i) RNA editing, (ii) alternative splicing and (iii) translation regulation.16 Nishihara et al.17 also showed that a very conserved sequence belonging to a SINE subfamily can act as a distal cis-regulatory element. The latter is a DNA sequence regulating the expression of a gene located on the same DNA molecule (hence the cis = intra-molecular) but is remote to the gene it affects. Such regulatory elements are often sites where trans-acting factors (intermolecular proteins) bind.
|
HERV Family |
Gene involved |
Function |
|
HERV-E18
|
Human apolipoprotein C1 (APOC1) gene |
Lipid metabolism |
|
HERV-E19 (alternative promoter)
|
Human endothelin B receptor (EDNRB) gene (on chromosome 13) |
The gene codes for the endothelin receptor type B (a 7-transmembrane receptor that mediates the vasoconstrictor actions of endothelins |
|
HERV-L20 |
Human beta 1,3-galactosyltrasferase 5 gene |
in type 1 Lewis antigen synthesis GIT and mammary gland
|
HERV-H21 |
HHLA2 and HHLA3 |
immuno-protection in intestinal tissues, kidney, and lung |
Table 10: Cases where HERV LTRs contribute to Gene Expression
(v) Cell metabolism
Rubin et al.22 and Liu et al.23 demonstrated that certain TE transcripts, specifically Alu RNAs, show a transient increase in the cytosol under numerous stressful states (e.g. viral infections, heat shock and exposure to cycloheximide). They proposed that these Alu RNAs might function in cell metabolism under stressful conditions.
(vi) Protection against some exogenous retrovirus infections
HERVs have also been shown to confer host cell resistance to some exogenous retroviruses by blocking the exogenous retrovirus replication by either receptor interference or through antisense mRNA.24
(to be continued)
References
- Zamudio, Bourc’his D. (2010), Transposbale Elements In The Mammalian Germiline: A Comfortable Niche Or A Deadly Trap? Heredity105: 92–104.
- Aravin A. A., Klenov M. S., Vagin V. V., Bantignies F., Cavalli, G., Gvozdev V. A. (2004), Dissection Of A Natural RNA Silencing Process In The Drosophila Melanogaster Germ Line. Mol. Cell. Biol. 24: 6742–6750.
- Chapman E. J., Carrington, J. C. (2007) Specialization And Evolution Of Endogenous Small RNA Pathways. Nat. Rev. Genet. 8: 884–896.
- Ding S. W. and Voinnet O. (2007), Antiviral Immunity Directed By Small RNAs. Cell 130-413.
- Hartig J. V. Tomari Y., Forstemann K. (2007), piRNAs The Ancient Hunters Of Genome Invaders. Gene Dev. 21: 1707–1713.
- Gasior S. L., Roy-Engel A. M., Deininger P. L. (2008), ERCC1/XPF Limits L1 Retrotransposition. DNA Repair (Amst).7(6): 983-9.
- Shi X., Seluanov A., Gorbunova V. (2006), Cell Divisions Are Required For L1 Retrotransposition. Mol. Cell. Biol. 27(4): 1264-1270.
8. Yoder J. A., Walsh C. P., Bestor T. H. (1997), Cytosine Methylation And The Ecology Of Intragenomic Parasites. Trends Genet. 13(8): 335-40.
- Medstrand P., Mager D. L. (1998), Human-Specific Integrations of the HERV-K Endogenous Retrovirus Family. J Virol.72(12): 9782–9787.
- Hughes J. F., Coffin J. M. (2001), Evidence For Genomic Rearrangements Mediated By Human Endogenous Retroviruses During Primate Evolution. Nature Genetics 29: 487 – 489.
11. Sha M., Xinhua L., Xiang-Ping L., Geertruida M. V., Finnerty H., Racie L., LaVallie E., Xiang-Yang T., Edouard P., Howes S., Keith J. C., McCoy J. M. (2000), Syncytin Is A Captive Retroviral Envelope Protein Involved In Human Placental Morphogenesis. Nature 403: 785-789.
- Kapitonov V. V., Jurka J. (2005), RAG1 Core And V(D)J Recombination Signal Sequences Were Derived From Transib Transposons. PLoS Biol3(6): e181.
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2002), Molecular Biology Of The Cell, Garland Science.
- Tarlinton R. E., Meers J., Young P. R. (2006), Retroviral Invasion Of The Koala Genome. Nature. 6; 442(7098): 79-81.
- Cooper G. M., Hausman R. E. (2006), The Cell: A Molecular Approach, Sinauer Associates, Inc.
- HaslerJ.,Strub K. (2006), Alu RNP And Alu RNA Regulate Translation Initiation In Vitro. Nucleic Acids Res. 34: 2374-2385.
- Nishihara H., Smit A. F., Okada N. (2006), Functional Noncoding Sequences Derived From SINEs In The Mammalian Genome. Genome Res.16: 864-874.
- Medstrand P., Landry J. R., Mager D. L. (2001), Long Terminal Repeats Are Used As Alternative Promoters For The Endothelin B Receptor And Apolipoprotein C-I Genes In Humans. J Biol Chem. 19; 276(3): 1896-903.
- Landry J. R., Mager D. L. (2003), Functional Analysis Of The Endogenous Retroviral Promoter Of The Human Endothelin B Receptor Gene. J Virol. 77(13): 7459-66.
- Dunn C. A., Medstrand P., Landry J. R., Mager D. L. (2003), An Endogenous Retroviral LTR Is The Dominant Promoter For Human Beta1,3-Galatosyltransferase 5 In The Colon. Proc Natl Acad Sci USA. 28; 100(22): 12841-6.
21. Mager D. L., Hunter D. G., Schertzer M., Freeman J. D. (1999), Endogenous Retroviruses Provide The Primary Polyadenylation Signal For Two New Human Genes (HHLA2 And HHLA3). Genomics. 1; 59(3): 255-63.
- Rubin C. M., Kimura R. H., Schmid C. W. (2002), Selective Stimulation Of Translational Expression By Alu RNA. Nucleic Acids Res. 30 (14): 3253-3261.
- Liu W. M., Chu W. M., Choudary P. V., Schmid C. W. (1995), Cell Stress And Translational Inhibitors Transiently Increase The Abundance Of Mammalian SINE Transcripts. Nucleic Acids Res. 23: 1758-1765.
24. Lankenau D. H., Volff J. N. (2009), Transposons and the Dynamic Genome, Springer

